Holy Grail of sugar biology
ETH-Zurich biologists have determined the structure of a bacterial enzyme that attaches sugars to proteins. The findings on the structure provide insights into the mechanism of this kind of protein modification. Furthermore, they can now develop new methods for the production of therapeutically useful “sweet proteins”.
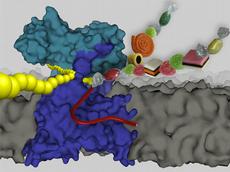
The enzyme of which the ETH-Zurich biologists have just determined the structure has an almost unpronounceable name: oligosaccharyltransferase. This protein sits in the membranes of certain bacteria, gram-negative bacteria, including various species of pathogenic campylobacter.
Thanks to this enzyme, these bacteria have a special ability that is otherwise restricted to eukaryotic cells like yeast or mammalian cells: they can attach a variety of sugars to proteins. This process is known as glycosylation and takes place at a particular part of the protein, namely an amino acid called asparagine. The enzyme can bind sugars to this component via a nitrogen group, which is why the process is also known as N-linked glycosylation.
Structural clarification first
Biologists – and chemists – have puzzled long and hard about how exactly this happens. After all, the reaction is inert as the nitrogen residue on the asparagine is not reactive enough. “The crux was how asparagine is activated to react with the sugar,” says Christian Lizak, a doctoral student at the Institute of Microbiology at ETH Zurich.
In order to chemically understand this elusive process, it was necessary to know the precise structure of the enzyme. Consequently, the researchers from ETH Zurich isolated and crystallised PglB from the bacterium Campylobacter lari. “This step was particularly challenging,” says Lizak, who is the first author of the study that has just been published in the journal Nature; “we started working on it eight years ago.”
The crystals were shot with X-rays at a high-resolution radiation source of the Swiss Light Source (SLS) at the Paul Scherrer Institute. Based on the obtained X-ray images, the researchers were able to calculate every single atom position with a resolution of 3.4 angstrom, which corresponds to a scale of a few billionths of a metre.
Vital function
In bacteria, the enzyme PglB is responsible for N-linked glycosylation. Compared to the enzyme in animal cells, consisting of eight different subunits, this oligosaccharyltransferase has a relatively simple structure. However, the central unit of the animal enzyme is very similar to the PglB protein of bacteria.
Even though N-linked glycosylation is not essential for bacteria, cells of higher organisms (from yeast up to mammals) cannot live without it: if this machinery is deleted in yeast cells, for instance, the cells die within a short period of time since N-glycosylation is important for a multitude of cellular functions. Over half of all proteins in a eukaryotic cell are “sweetened” with sugars after synthesis.
“The structure of the oligosaccharyltransferase is something like the ‘Holy Grail’ of glycobiology,” says Lizak. Several of the world’s leading research teams from Oxford, Caltech or MIT have tried in vain to solve its structure. The researchers from ETH Zurich have now managed to take a big step forward. “The basis for our success was the collaboration of different competence centres within ETH Zurich,” stresses the doctoral student.
The original system for N-linked glycosylation in bacteria comes from ETH-Zurich professor Markus Aebi’s lab, where Lizak did his doctoral thesis and learned the secrets of sugar biology. When he hit a brick wall in the crystallisation of the enzyme, he was able to switch to Kaspar Locher’s team during his dissertation. Locher is an expert in purifying, crystallising and solving the structure of membrane proteins, and PglB belongs to this class of proteins. In this way it became possible to crystallise the enzyme. Using the obtained X-ray data, the professor for molecular membrane biology from ETH Zurich was able to determine the three-dimensional structure of the protein.
Tool for production of therapeutics
The enzyme structure provides fundamental insights into understanding the process of N-linked glycosylation both in bacteria and eukaryotes. The atomic composition and three-dimensional shape reveal how the enzyme recognises its substrates, where and how it binds them and how it attaches the sugar to different proteins.
This result now gives the researchers a tool to specifically modify the enzyme. The “normal” enzyme can only glycosylate particular proteins having a specific sequence of amino acids. However, with a modified enzyme other substrates could also fit into its active domain and could get glycosylated.
If the researchers succeed in converting the enzyme, they could use it in E. coli bacteria to produce sweetened proteins that can be used as therapeutic agents or vaccines.
“Like DNA and proteins, sugars encode biological information and represent the third language of life. Through the structural determination of the oligosaccharyltransferase, we know now how this information carrier can find its way onto proteins,” says Lizak.
Further reading
Lizak C, Gerber S, Numao S, Aebi M & Locher KL. X-ray structure of a bacterial oligosaccharyltransferase. Nature online publication, 15. Juni 2011. DOI: 10.1038/nature10151







READER COMMENTS